By Molnár-Institute for…
Creating QbD multifactorial design spaces using MOLNÁR-INSTITUTE’S DryLab®4 Robustness Module
Three studies indicate the usefulness of the Robustness module built into Molnár-Institute’s proprietary software suite, DryLab®4, in facilitating easier regulatory submissions that meet recent demands for managing a method-life-cycle underpinned by a Quality-by-Design (QbD) model.
Since 2002, when the US Food and Drug Administration (FDA) launched its “Pharmaceutical Current Good Manufacturing Practices (cGMPs) for the 21st Century” initiative, QbD approaches have become increasingly essential in pharmaceutical development.
The subsequent International Council for Harmonization (ICH) quality guideline Q8 (R2) recommends that an enhanced, quality by design approach to product development would be based on ‘’systematic evaluation, understanding and refining of the formulation and manufacturing process’’, that would include the enhanced product and process understanding in combination with quality risk management to establish an appropriate control strategy, which can, for example, include a proposal for a design space and/or real-time release testing. This more systematic approach could facilitate continual improvement and innovation throughout the product lifecycle, as demanded by ICH Q10).
Elimination of human mistakes is one of the major drivers behind current regulatory approaches, also typified by the FDA’s Data Integrity and Analytical Quality by Design (AQbD) guidelines.
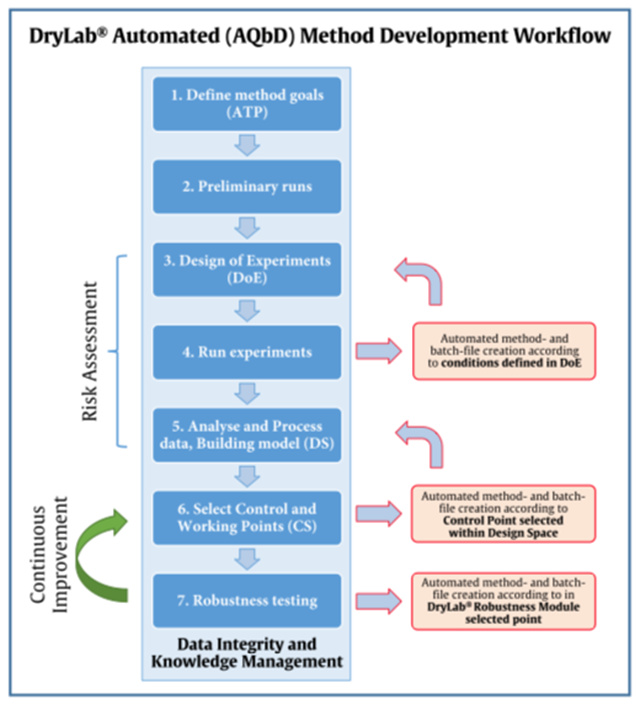
The QbD and AQbD concepts can be extended and allied to DryLab®4 automation to create systematic yet speedy method approaches that include defining methods goal, risk assessment, developing a Design Space, implementing a control strategy and continual improvement to increase method robustness and knowledge.
Creating a QbD design space
The first featured study, from 2013, was conducted by Dr. Alexander H. Schmidt of Steiner & Co., Deutsche Arzneimittel GmbH, and Molnár-Institute President, Dr. Imre Molnár. They examined ‘’Using an innovative Quality-by-Design approach for development of a stability indicating UHPLC method for ebastine in the API and pharmaceutical formulations’’ [1]. This sought to improve on the conventional practice of developing an analytical method in liquid chromatography using trial- and-error, in varying one-factor-at-a-time (OFAT), examining resolution of peaks to identify the best method. This time-consuming approach required intensive manual data interpretation and often resulted in a non-robust performance, when transferred into another lab, because interactions between factors were not considered.
The first in-silico retention modelling initiative, based on solid science, was pioneered by Lloyd Snyder and co-workers from 1986, giving birth to the “DryLab®” software to replace OFAT approaches for first isocratic methods [2] and subsequently for gradient installations [3].
The Schmidt and Molnár study, published in The Journal of Pharmaceutical and Biomedical Analysis, aimed to achieve successful chromatographic separation of the API ebastine from impurities using a Waters Acquity UPLC BEH C18, 50mm×2.1mm, 1.7 µm particle size column with gradient elution of 10mM acetate buffer pH 6.2 and a mixture of acetonitrile/2-propanol (1:1) as the mobile phase. This separation was the platform for incorporating QbD principles to the method development approach by using the chromatography modeling software DryLab®4 to visualize a “Design Space” region in which changes to method parameters would not significantly affect the results as defined by ICH Q8.
DryLab®4 was used for screening and optimization of gradient time, temperature, solvent composition and pH to separate a mixture of ebastine and seven impurities. The software package included the PeakMatch and 3-D-Robustness modules, with statistical analysis calculated using MVA statistical software from Novia.
The robustness of the developed method was studied by varying the six parameters: gradient time, temperature, ternary composition of the eluent, flow rate and start and end concentration of the organic eluent at 3 levels (+1, 0, −1). The resulting 729 experiments were performed in silico within DryLab®4 from the previously constructed model for Design Space and they showed that the required resolution of 2.0 was attainable in all experiments. To prove the stability-indicating performance of the method, forced degradation (acid and base hydrolysis, oxidation, photolytic and thermal stress conditions) of ebastine was carried out. Baseline separation could be achieved for all peaks of the impurities, the degradation products and the API. Total run time was only 4 minutes, representing a 40-fold increase in productivity in comparison to the method published in the Ph. Eur. monograph, allowing purity testing of more than 360 samples per day.
A verification study demonstrated the accuracy of the established Design Space model with a relative error of prediction of only 0.6%. The method was further validated for specificity, linearity, accuracy and precision, and robustness in compliance with ICH guideline Q2 (R1). The method was found to be linear in the concentration range from the quantification limit (LOQ) to 125% of the specification limit for ebastine and each of the impurities with correlation coefficients of not less than 0.999. The recovery rate was between 98.15 and 100.30% for each impurity. The repeatability and intermediate precision (RSD) were less than 3.2% for ebastine and each of the impurities.
Building a multidimensional design space
The same two authors were joined by Mijo Stanic, also from Steiner & Co, for a subsequent study ‘‘In silico robustness testing of a compendial HPLC purity method by using of a multidimensional design space build by chromatography modelling—Case study pramipexole’’ that was published in the JBPA in 2014[4].
Here, the study team sought to impose a QbD approach on purity testing of the API pramipexole performed using the official (compendial) and harmonized method published in the European Pharmacopeia (Eu.Ph.) and United States Pharmacopeia (USP), which demands use of a C18 column with gradient elution of an ion-pairing-buffer of pH 3.0 (mobile phase A) and acetonitrile (mobile phase B).
Although not recommended in general, compendial methods are often adapted for purity testing of generic formulations. This study describes a novel approach to evaluate method robustness of an adapted method, prior to full method validation. Based on QbD principles, they performed a small number of experiments, entering them into the DryLab®4 software, including PeakMatch and 3-D-Robustness modules, to allow visualization of a multidimensional “Design Space”, a region in which changes in method parameters will not significantly affect the results, as defined in the ICH guideline Q8 (R2) leading to a more flexible method handling in routine analysis.
For two different recommended C18 columns, the team constructed a multidimensional Design Space (Method Operating Design Region, MODR) to study the robustness of the adapted method with the DryLab®4 Robustness Module, using retention times and peak areas of individual peaks from the experimental runs as input data for HPLC modeling.
For the separate Eluent Design Space study DryLab®4 was used, for which Molnár-Institute constructed a specific ionic strength vs. ion-pairing concentration model.
In a full factorial design, the following six parameters were varied at three levels (low, nominal, high): gradient time, temperature, pH of the aqueous eluent (A), flow rate, start- and end concentration of the organic mobile phase component (eluent B). The resulting 36 = 729 experiments were performed in silico from the previously constructed models for Design Space in less than one minute and showed that the required resolution of 2.0 could not be reached in all experiments for the two columns as recommended by Eu.Ph., with failure rates of 25% and 16%, respectively. However, by adjusting gradient time, the team was able to fulfill the requirements with a failure rate of zero.
The team thus succeeded in their goals of establishing a multi-factorial Design Space that could be used for in silico robustness testing, as well as increasing flexibility of the method handling in the routine work, thus reducing overall process development time from months to days.
Automated separation of 10 compounds using DryLab®4
In 2018, Arnold Zöldhegyi led a team representing the Molnar-Institute and Moscow’s Mendeleev University of Chemical Technology in a study of ‘‘Automated UHPLC separation of 10 pharmaceutical compounds using software-modeling’’[5].
This work used the DryLab®4 automation tool to perform Design of Experiments (DoE) runs, also with Design Space verification runs and robustness calculation confirmation. Their whole development procedure followed an Analytical Quality by Design (AQbD) workflow and was documented accordingly. Error-proof method modelling again enabled rapid development with zero defects and, when compared, simulated and experimental results were found to be in very close alignment.
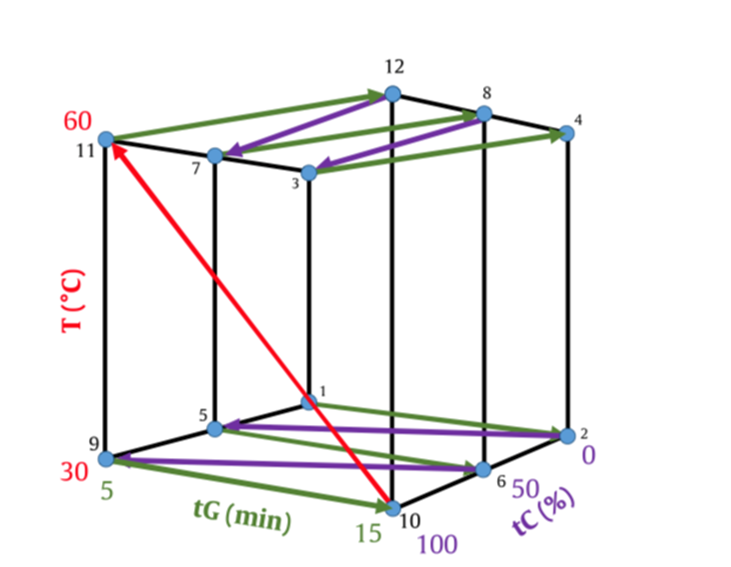
Automation routines can reduce human error, increase productivity and reduce per unit costs. Therefore, in this project, the goal was to develop an automation feature in order to reduce the time to create the necessary experiments, along with the time required to manually enter the run parameters. To achieve this goal, the team created a model mixture of blockbuster drug components to be able to follow a large number of experiments and test the influence of variabilities needed to program the automation algorithms in robustness studies and to control the experimental outcome of the individual separation conditions. This would eliminate possible errors on setting the conditions for each single run. The automated conduction of 12 numbered runs could then be imported into DryLab®4 and consequently add to proficient pharmaceutical Quality Control.
Furthermore, the team aimed to find an adequate method that could deliver an Analytical Target Profile (ATP), in this case based on resolution for the critical peak pair, that would provide a baseline, even when variables were altered between their tolerance limits.
To develop smooth, fast and robust methods free of human failures, the team developed a state-of-the-art automation method, using DryLab®4 to analyze data from a model mixture of ten drugs subjected to testing. Instrumental setup and data collection were performed with Shimadzu Nexera X2 and LabSolutions (Version 5.89), while DryLab®4 handled data processing and automation protocol.
The creation of this kind of modelling is most efficient, due to the simple measurement of peak positions and their movement caused by alterations of the most important parameters such as tG, T, pH, ternary composition (tC) or additive concentration (aC). With data generated out of only 12 input experiments, once the peak-table is matched, DryLab®4 can create multi- dimensional resolution models that can reliably predict critical resolution and retention times for millions of chromatograms. Among those, the best robust separation conditions can be easily selected within seconds.
Following AQbD-principles, the best available working point was selected and conformational experimental runs performed using the six worst cases of the conducted robustness calculation. Out of the 729 experiments modelled in DryLab®4’s robustness module, the worst cases were those most likely to fail if performed under varying, in-vivo lab conditions. Automation was executed to confirm those experimental runs as well. Once again, the automated results showed an excellent agreement with values predicted in simulation. This proved the usefulness and effectiveness of an automated, software-assisted analytical method development.
Correct matching of corresponding peaks under different conditions is often the Achilles-heel of model creation. Peak tracking in DryLab®4 bases on the assumption that while the same sample amount was injected all the time, after each run, single and total peak areas remain closely similar. As it was presented by Kormány et al. (2013), in most cases this offers a transparent way of following peak-movements [6]. In some cases, however, especially for close-eluting and for compounds present in similar amount, peak- tracking becomes difficult and less obvious. If the model turns out to show low predictive validity, troubleshooting has to be performed on previous steps.
Fortunately, similarly to DoE-runs, in DryLab®4 these runs can be carried out also in an automated way.
References
- Alexander H. Schmid, A.H., Molnár, I., (2013) ‘Using an innovative Quality-by-Design approach for development of a stability indicating UHPLC method for ebastine in the API and pharmaceutical formulations’; Journal of Pharmaceutical and Biomedical Analysis, 78–79, pp. 65–74. DOI: http://dx.doi.org/10.1016/j.jpba.2013.01.032
- Snyder, L.R, Dolan, J.W., Lommen, D., (1989) ‘DryLab® computer simulation for HPLC method development I. Isocratic elution’, Chromatogr. 485 (1989) 65–89. DOI: http://dx. doi.org/10.1016/S0021-9673(01)89133-0.
- Dolan, J.W., Lommen, D., Snyder, L.R., (1989), ‘DryLab® computer simulation for HPLC method development II. Gradient elution’, Chromatogr. 485 (1989) 91–112. DO!: http:// dx.doi.org/10.1016/S0021-9673(01)89134-2.
- Alexander H. Schmid, A.H., Molnár, I., Stanic, M., (2014) ‘In silico robustness testing of a compendial HPLC purity method by using of a multidimensional design space build by chromatography modelling—Case study pramipexole’; Journal of Pharmaceutical and Biomedical Analysis, 91, pp. 97–107. DOI: http://dx.doi.org/10.1016/j.jpba.2013.12.023
- Zöldhegyi, A., Rieger, H-J., Molnár, I., Fekhretdinova, L. (2018) ‘Automated UHPLC separation of 10 pharmaceutical compounds using software-modeling’; Journal of Pharmaceutical and Biomedical Analysis, 156, pp. 379–388. DOI: https://doi.org/10.1016/j.jpba.2018.03.039
- Kormány, R., Molnár, I., Rieger, H-J. (2013) ‘Exploring better column selectivity choices in ultra-high-performance liquid chromatography using Quality by Design principles’, Journal of Pharmaceutical and Biomedical Analysis 80, pp. 79–88. DOI: http://dx.doi.org/10.1016/j.jpba.2013.02.028
Resources
Click on Schmidt and Molnar (2013) for details of Study 1.
Click on Schmidt et al. (2014) for details of Study 4.
Click on A. Zöldhegyi et al. (2018) for details of Study 5.