By Molnár-Institute for…
Separation and characterization of proteins using DryLab®4 based software modeling
Molnár-Institute’s proprietary software suite, DryLab®4, provides analytical scientists with the tools needed to achieve separation and detailed characterization of therapeutic proteins biopolymers and antibody drug conjugates (ADCs) using liquid chromatography.
Application of DryLab® modelling in separation and characterization workflows is highlighted in four studies, all published in the Journal of Pharmaceutical and Biomedical Analysis.
Separating mAbs and ADCs
The first two studies, ‘’Practical method development for the separation of monoclonal antibodies and antibody-drug-conjugate species in hydrophobic interaction chromatography, part 1: optimization of the mobile phase’’[1] and ‘’Practical method development for the separation of monoclonal antibodies and antibody-drug-conjugate species in hydrophobic interaction chromatography, part 2: Optimization of the phase system’’[2], examine practical method development for the separation of monoclonal antibodies and antibody-drug-conjugate species in hydrophobic interaction chromatography, with Part 1 dealing with optimization of the mobile phase and Part 2 detailing optimization of the phase system.
In the Part 1 study [1], lead authored by Dr. Marta Rodriguez-Aller, of the School of Pharmaceutical Sciences, Universities of Geneva/Lausanne, the study team developed recommendations for method development in hydrophobic interaction chromatography (HIC) using monoclonal antibodies (mAbs) and antibody-drug conjugates (ADCs) as model drug candidates. They also examined HIC method development using DryLab® as optimization software.
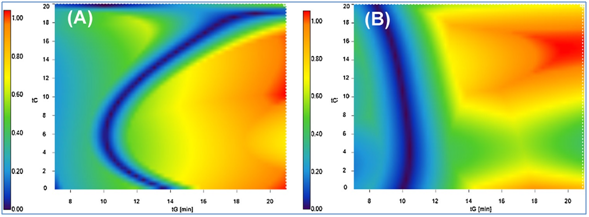
The study team found that gradient steepness played a major role for tuning retention and selectivity and should be optimized both for mAbs and ADCs. They highlighted possible addition of organic modifier (i.e. isopropanol) to the mobile phase in HIC, as particularly useful for mAbs separations. However, addition of isopropanol was never beneficial, when dealing with the analysis of ADC DAR (Drug-Antibody-Ratio) species in HIC, since the most hydrophobic species (DAR6 and DAR8) cannot be eluted in the presence of ∼10% isopropanol.
They succeeded in developing HIC automated method optimization procedure using DryLab® software, achieving low average retention time errors between predicted and experimental retention times of around 1% for an optimization procedure, that took only one hour to process two gradients for each sample and equilibration.
Optimizing the phase system
The Part 2 study, examining optimization of the phase system in the separation of mAbs and ADCs, was led authored by Dr. Alessandra Cusumano, of the University of Bologna’s Alma Mater Studiorum Department of Pharmacy and Biotechnology [2]. This study set out to evaluate the performance of commercially available HIC columns and develop a fast and automated “phase system” (i.e. stationary phase and salt type) optimization procedure for the analytical characterization of protein biopharmaceuticals. The team selected various therapeutic mAbs (denosumab, palivizumab, pertuzumab, rituximab and bevacizumab) and a cysteine linked ADC (brentuximab-vedotin) as model substances and evaluated several HIC column chemistries (butyl, ether and alkylamide) from different providers in four different buffer systems (sodium acetate, sodium chloride, ammonium acetate and ammonium sulfate).
Again, they used DryLab® computer assisted retention modeling to achieve high throughput optimization of separations after selection of the most appropriate phase systems.
The study found historical TSK gel Butyl NPR phase and the brand new Thermo MAbPac HIC-10 to be the most versatile stationary phases for hydrophobicity, peak capacity and achievable selectivity, with ammonium sulfate and sodium acetate salt types found to be particularly well adapted for analytical characterization of mAbs and ADCs, given higher concentrations of sodium acetate versus ammonium sulfate to achieve similar retention in HIC.
Analytical QbD platform
The third study, presenting A platform analytical quality by design (AQbD) approach for multiple UHPLC-UV and UHPLC–MS methods development for protein analysis, was produced by a team from the Analytical Science and Technology, Global Manufacturing Science and Technology, at Sanofi, in Framlingham MA, headed by Dr. Jianmei Kochling, and published in March 2016 [3].
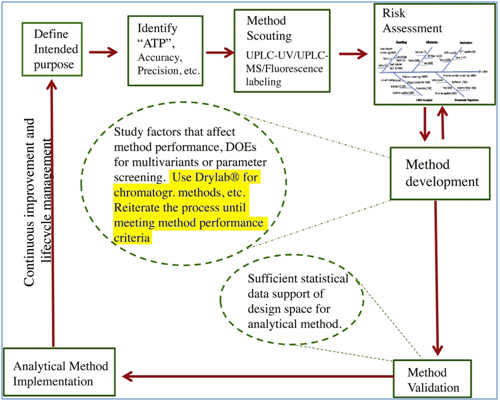
The team sought to develop a platform analytical Quality by Design (QbD) approach for methods development that could also be exploited across a range of applications in methods development that shared common equipment and procedures.
Based on a development process using three methods, they proposed a systematic approach strategy, in which knowledge gained from the UHPLC-UV peptide mapping method could be easily transferred to UHPLC–MS oxidation or UHPLC-UV C-terminal heterogeneity for the same protein. In addition, their platform AQbD method development strategy ensured method robustness was built in during development.
The study highlighted several key advantages for following the AQbD approach, including generating reliable data for product development, avoidance of extensive post-approval analytical method change and more timely data release, reduced regulatory risk, and lowered lab operational cost in the commercial phase. Moreover, large, reliable database and knowledge gained during AQbD method development could provide strong justifications during regulatory filling for the selection of important parameters or parameter change needs for method validation, and help to justify for removal of unnecessary tests used for product specifications.
RPLC separation of ADCs
The most recent study, Separation of antibody drug conjugate species by RPLC: A generic method development approach, was published in January 2017 and was led authored by Prof. Szabolcs Fekete of the Universities of Geneva/Lausanne School of Pharmaceutical Sciences, assisted by the School’s Dr. Davy Guillarme and Molnár-Institute founder and President, Dr. Imre Molnár [4].
This study examined the use of DryLab® modelling software for the successful method development of IgG1 cysteine conjugated antibody drug conjugate (ADC) in reverse phase liquid chromatography (RPLC). The ultimate goal was to be able to calculate the average drug to antibody ratio (DAR) of any ADC product.
The team proposed a generic method development strategy, including optimization of mobile phase temperature, gradient profile and mobile phase ternary composition. The study pioneered use of DryLab®’s 3D retention modelling capabilities for a large therapeutic protein. Based on a limited number of preliminary experiments, the team achieved a fast and efficient separation of the DAR species of a commercial ADC sample (brentuximab vedotin), with retention model prediction that was highly reliable, with average error of retention time prediction below 0.5% using either 2D or 3D models.
Up to six initial experiments were required to build the 2D retention models, while 12 experiments were recommended to create the 3D model.
The study concluded that RPLC could now be considered as a good method for estimating the average DAR of an ADC, based on the observed peak area ratios of RPLC chromatogram of the reduced ADC sample. Furthermore, by understanding the retention behavior of ADC peaks, overall analysis time could be reduced from more than an hour to as little as 20 minutes.
Study References
- Rodriguez-Aller, M., Guillarme, D., Beck, A., and Fekete, S. (2016) ‘Practical method development for the separation of monoclonal antibodies and antibody-drug-conjugate species in hydrophobic interaction chromatography, part 1: optimization of the mobile phase’, J Pharm Biomed Anal, 118, pp. 393-403. https://doi.org/10.1016/j.jpba.2015.11.011
- Cusumano, A., Guillarme, D., Beck, A., and Fekete, S. (2016) ‘Practical method development for the separation of monoclonal antibodies and antibody-drug-conjugate species in hydrophobic interaction chromatography, part 2: Optimization of the phase system’, J Pharm Biomed Anal, 121, pp. 161-173. https://doi.org/10.1016/j.jpba.2016.01.037
- Kochling, J., Wei, W., Yimin, H., Qian, G., and Castaneda-Merced, J., (2016) ‘A platform analytical quality by design (AQbD) approach for multiple UHPLC-UV and UHPLC–MS methods development for protein analysis’, J Pharm Biomed Anal, 125, pp. 130-139. https://doi.org/10.1016/j.jpba.2016.03.031
- Fekete, S., Molnár, I., Guillarme, D., (2017) ‘Separation of antibody drug conjugate species by RPLC: A generic method development approach’, J Pharm Biomed Anal, 137, pp. 160-169. https://doi.org/10.1016/j.jpba.2017.01.013