By Molnár-Institute for…
Combination method for ion exchange (IEX) separations of MAbs
Berlin: – An innovative methodology to speed up the chromatographic characterization of anti-COVID-19 protein therapeutics using a combination of automated, systematic modeling and ultra-short liquid chromatography (LC) columns, co-pioneered by Molnár-Institute for Applied Chromatography has shown application to ion exchange (IEX) chromatographic separations.
The study has yielded highly encouraging results across a range of chromatographic techniques, including IEX.
The study, ‘Expediting the chromatographic analysis of COVID-19 antibody therapeutics with ultra-short columns, retention modeling and automated method development’ is published in the Journal of Pharmaceutical and Biomedical Analysis .[1].
Response to COVID-19
The research, led by Bastiaan Duivelshof from the University of Geneva, also involved Molnár-Institute application specialist Arnold Zöldhegyi and Waters Corporation scientists Szabolcs Fekete and Matthew Lauber. It was launched as a joint response to the need exposed by the COVID-19 pandemic to accelerate emergency use authorization (EUA) of new therapeutics and vaccines that included several monoclonal antibodies (mAbs).
Since EUA submissions usually require scientific data on drug candidates to be collated in parallel with drug development, there is little time to spare for misguided method development or waiting on traditional turnaround times for LC characterizations.
The team took this as their cue to investigate new means of expediting rapid chromatographic characterization of protein therapeutics that combined the use of automated, systematic modeling and new ultrashort column designs to quickly optimize high throughput separations for 2 exemplar COVID-19-related mAb products: casirivimab-imdevimab cocktail and tocilizumab.
Combination method
Among others, the study demonstrated that using the new combination method, both studied mAb-samples could be characterized by IEX analysis in less than two-minute method run time. This also means that in case of a common suite of four LC profiling techniques (RP, IEX, HILIC and SEC) could be realized in less than an one hour turnaround time.
Untapping the full resolving potential of the new column hardware and speeding up the entire analytical process with the aid of computerized modeling—Molnár-Institute’s DryLab® software platform—the number of wet-lab experiments could effectively be reduced and quickly complex chromatographic data contextualized in the acquired Design Spaces [2].
The other main innovation used in the study was a streamlined analytical quality-by-design (AQbD) modeling workflow based on automated generic approaches (Design-of-Experiments) for monoclonal antibody and antibody-drug-conjugate separations. This takes into account previous scientific results that reveal the propensity of large proteins to be extremely sensitive to the mobile phase composition, becoming eluted in many types of separations with only a very minor change in eluent strength (%B), referred to as the “on-off” phenomenon or “bind and elute” retention mechanism. The prevalence of this effect increases with protein molecular size (i.e. number of available charges) in IEX. A consequence of “on-off” elution behavior is that only the first segments of a column actively takes part in peak retention and separation, which also means very short (15–20 mm) columns can be used to make separations faster without compromising efficiency.
IEX results
Cation-exchange chromatography (CEX) is usually employed for charge variant analysis and to identify and quantify acidic versus basic species with two approaches often used to elute antibody species. Salt-gradient technique weakens the strength of ionic interactions between the analyte and the stationary phase by increasing the ionic strength of the mobile phase. Alternatively, pH-gradient separation changes the pH of the eluent over time to decrease the exposed charge of the solute and promote elution.
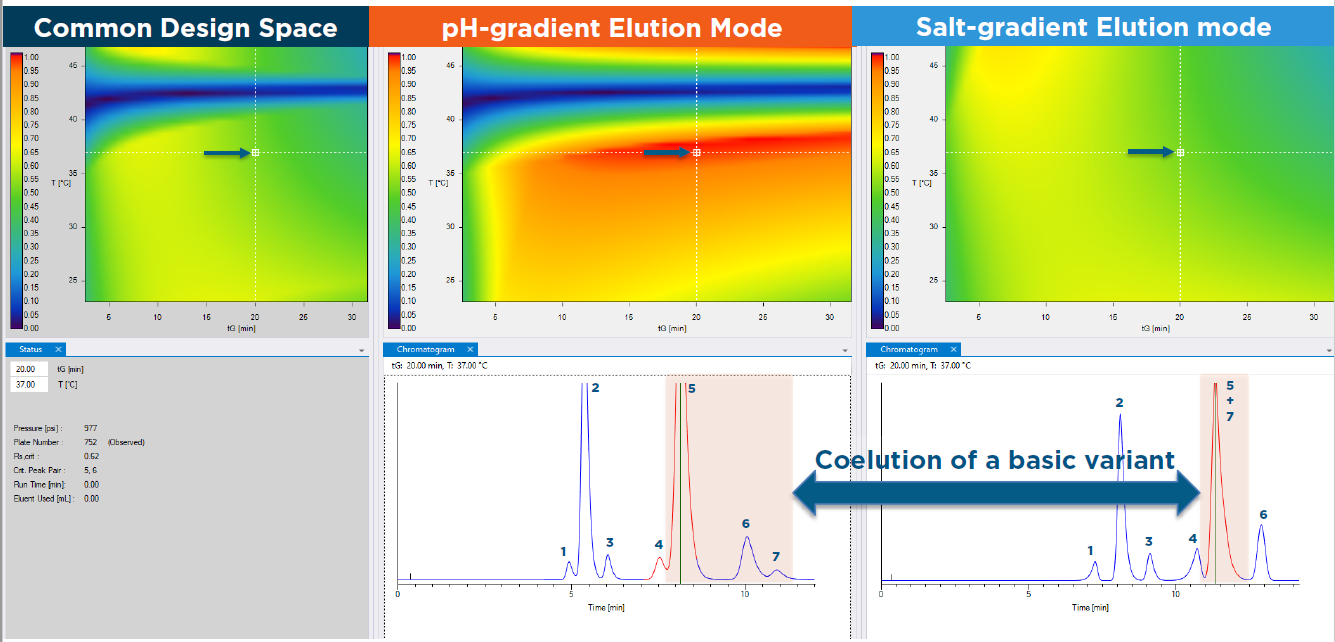
To discover underlying differences in the gradient modes, the team carried out systematic studies by performing 2D gradient time-temperature separation models for both intact casirivimab and imdevimab and their subunits. For a direct comparison, researchers applied the new DryLab® Software Design Space Comparison solution, originally designed for the comparison of complete separation systems, including separations obtained on different columns ran in a single type of separation mode. In this case however, design spaces were innovatively used to head-to-head compare columns operated in two substantially different elution modes (Figure 1 Model). It is well-known IEX separations differentiate on the basis of electrostatic interactions present between the exchanger-groups and accessible charges of large solutes, which is why baseline resolution is usually hard to achieve for many critical charge variants. In fact, shoulder peaks for acidic and basic variants are frequently encountered, but an Rs value of 1 is closer to what is practically achievable (in contrast to the Rs value of at least 1.5 specified in guidelines like the United States Pharmacopeia).
As it was shown, the resolution maps suggested that an overall higher critical resolution could be obtained with the pH gradient (Rs,crit = 0.98) versus the salt gradient technique (Rs,crit = 0.69). However, for the most part, the two modes provided fairly similar elution profiles, but with slightly better separations of basic species achieved with pH gradient elution, especially for the digested imdevimab sample. Therefore, the pH gradient mode was further optimized for each sample.
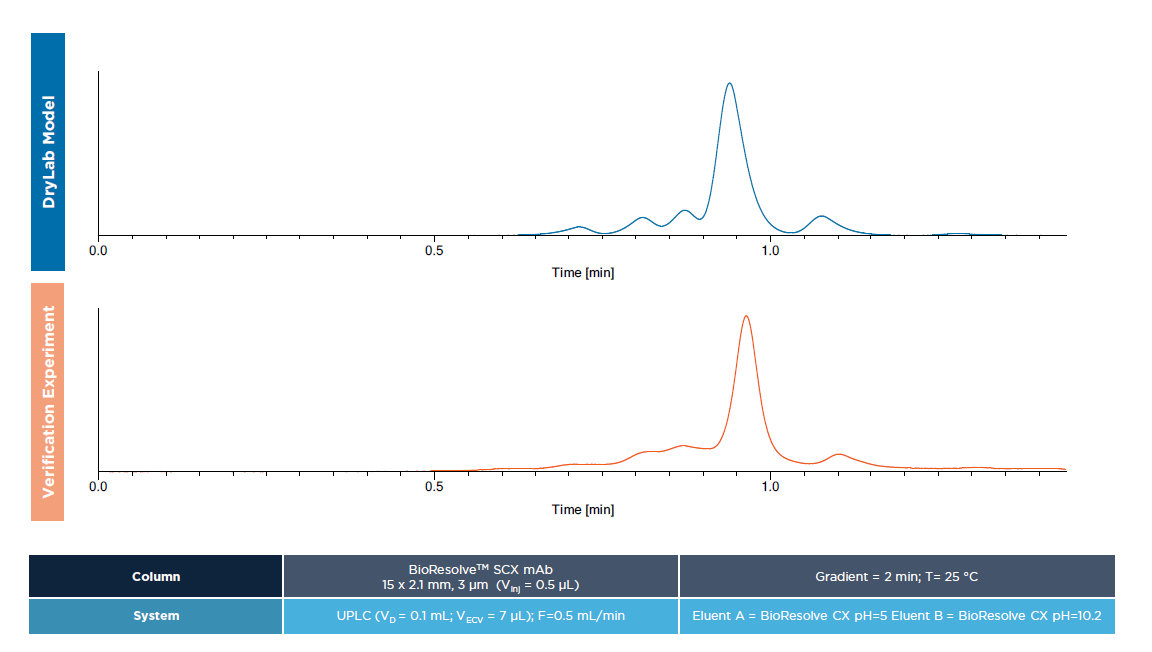
After finding the optimum conditions on commercial 2.1 × 50 mm columns, the methods were virtually transferred to prototype 2.1 × 15 mm ultra-short columns along with the employment of state-of-the-art low dispersion chromatographic system with a low gradient delay volume. A significant advantage of a virtual model transfer was that the complete systematic modeling context could be retained, which was ultimately used to further model-speed-up the analysis by decreasing the gradient time and applying a proportional increase of the flow rate. Finally, it was found that a tG = 2 min gradient at F = 0.5 mL/min flow rate performed well for all four samples using the following gradient programs: 10–30 %B for intact casirivimab, 0–25 %B for partially digested casirivimab, 15–35 %B for intact imdevimab and 5–35 %B for the partially digested imdevimab. Temperature was set at T = 25 °C since the model revealed the elevated temperature would not benefit to any substantial improvement in acquired resolutions. For the last, high predictive capability of the model was experimentally proven, the average error of retention time between the transferred model run and the corresponding experiment was found less than 3%.
FIGURE 2 (Verification)
Mab optimization within minutes
The team identifies practical benefits of combining retention modeling and automated experiments with use of short protein columns for mAb analysis.
It suggests that the whole process to develop and optimize four chromatographic methods (also including other important protein LC modes) for two mAb samples might require only two days of experimental work, compared with at least a week following a traditional methodology.
“Retention modeling enables the in silico optimization of mAb (or any protein) separations within a few minutes. The automation module of DryLab® Software significantly simplifies manual work and thus speeds up the method development procedure,” the study concludes.
By employing ultrashort protein columns (15–20 mm long for RPLC and IEX) of Waters, analysis times, as low as 1–2 min could be obtained, marking a significant time savings in comparison to todays industry practice, often having more than 20–30 min long methods.
“Overall, we have shown that conventional chromatographic techniques can quickly be transferred in silico to short columns, higher throughput parameters and to low dispersion chromatographic systems without the need for further experiments. These approaches might be valuable for turning around scientific data on drug candidates during critical times, when the preparation of filing information must be accelerated,” the study concludes.
References
- Duivelshof, B., Zöldhegyi, A., Guillarme, D., Lauber, M. and Fekete, Sz. (2022). Expediting the chromatographic analysis of COVID-19 antibody therapeutics with ultra-short columns, retention modeling and automated method development. Journal of Pharmaceutical and Biomedical Analysis, [online] 221, p.115039. doi:10.1016/j.jpba.2022.115039.
- den Uijl, M.J., Schoenmakers, P.J., Pirok, B.W.J. and Bommel, M.R. (2020). Recent applications of retention modelling in liquid chromatography. Journal of Separation Science, 44(1), pp.88–114. doi:10.1002/jssc.202000905.
- Fekete, S., Rudaz, S., Fekete, J. and Guillarme, D. (2012). Analysis of recombinant monoclonal antibodies by RPLC: Toward a generic method development approach. Journal of Pharmaceutical and Biomedical Analysis, 70, pp.158–168. doi:10.1016/j.jpba.2012.06.021.
About MOLNÁR-INSTITUTE
Founded in 1981, Molnár-Institute develops DryLab®4, a software for UHPLC modelling for a world-wide market. Its powerful modules gradient editor, peak tracking, automation, robustness and Design Space Comparison allow for the most sophisticated method development as required across modern pharma industries. Analytical scientists use DryLab®4 to understand chromatographic interactions, to reduce analysis time, to increase robustness, and to conform to AQbD-principles, according to the recently published ICH Q14 regulatory framework, where it has to be explained, why a certain setpoint was fixed at its selected location and not somewhere else.
The Molnár-Institute is a registered partner to the US-FDA, CDC and many other regulatory bodies. DryLab®4 has pioneered AQbD long before regulatory agencies across the world encouraged such submissions. Widely implemented by thought leaders, the software contributes substantially to the paradigm shift towards a science and risk driven perspective on HPLC Quality Control and Assurance.
Further information at: http://www.Molnár-institute.com/
Resources
Click on Expediting the chromatographic analysis of COVID-19 antibody therapeutics with ultra-short columns, retention modeling and automated method development (2022) to access full study.