By ATG:biosynthetics GmbH
ATG Eukaryotic Gene Expression Modulation via BioDesign
Overview
ATG:biosynthetics (ATG) offers Molecular BioDesign solutions that combine the proprietary EvoMAGIS (in silico evolution mediates to artificial genomes) bioinformatics platform with the realization of genes and clusters via gene syntheses.
BioDesigned synthetic eukaryotic genes are tailored to realize sequence modulated physical DNA-sequences like single genes or multiple formal-functional gene variants for gene activity analyses and minimal functional gene fragment libraries.
Modular BioDesign can be applied to epitope screening, biochemical gene clusters and hetero-protein complex coding gene clusters, combinatorial vector designs, gene libraries and a wide range of sequence calculations up to 16 kb, according to individual customer requirements.
All these different approaches can be mutually realized to fit particular experimental and developmental needs.
Background
In contrast to functional improvements of bacterial expression Eukaryotic expression optimization procedures need to have an impact on additional molecular levels and more functional variants are to be considered.
Basic cellular transcription is characterized by the principles of general transcription of housekeeping genes, cellular differentiation and its specific promoters with selective transcription factor binding sites in the genome. The cytosolic expression machine is using differentiation reflecting codon tables with specific variant codon frequency related to the frequency of altered amino-acyl-tRNA concentration. Especially for the substantial improvement of heterologous expression many more molecular parameters especially on the mRNA level and protein level need to be considered to approach a realistic view of functional expression. Cellular traffic for localization and export of proteins as well as its glycosylation are additional features that impact on the final outcome of the correct function in terms of yield of mature vs. premature product accumulation as well as the desired maximum specific activity of the final gene products.
Modular BioDesign
ATG:biosynthetics has a strong background in comparative genomics bioinformatics, a source of valuable information to access, that is highly supportive for ATG’s BioDesigns. For functional expression parameter analyses, ATG can call on long experience in gene services and excellent proprietary bioinformatic tools to provide competitive advantage.
ATG BioDesigned synthetic genes are tailored for the realization of sequence modulated physical DNA-sequences like single genes, multiple formal-functional gene variants for gene activity analyses and minimal functional gene fragment libraries. Applications of Genomics based BioDesigns in this field are calculating high density peptide microarrays for epitope screening, biochemical gene clusters and hetero-protein complex coding gene clusters, combinatorial vector designs, gene libraries and all different kind of sequence calculations according to customer requirement specifications (up to 16 kb).
All these different approaches can be realized in a mutual realization process with the customers to meet individual experimental and developmental requirements.
Working from available genomic data
ATG integrates the highest level of molecular data sets available and genomic analyses as well as the related literature. Condensing all this information provides valuable decision process relevant information about the maximum formal- functional molecular requirements. Pareto optimized sequence designs are realized by simultaneous multi-parametric sequence calculations. This allows the analysis to achieve the best functional tradeoffs between sequence requirement constraints.
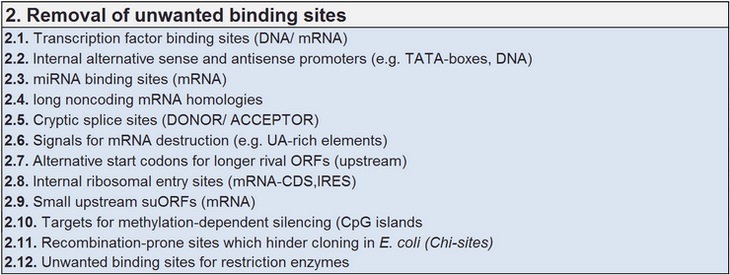
Removing unwanted binding sites and sequence motifs
Sequence motifs are either desired or need to be identified and eliminated in case they occur arbitrarily during the sequence modulation processes in statu nascendi. Many of the functional sequence motifs show a degenerate sequence composition, resulting in variations that can be described in base weighted libraries. Desired motifs can be introduced during the multi-parametric sequence modulation processes while modulating other sequence parameters like the codon use.
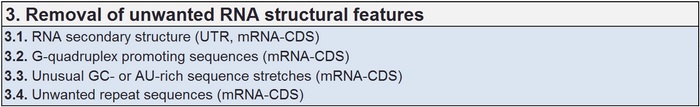
Removing unwanted mRNA structural features
Smooth transcripton and translation processes are dependent on the mRNA structure but pausing signals can also be highly valuable in assuring a functional outcome. Many of these RNA bound expression relevant parameters can be analysed in detail and considered in the final design before starting any experiments.
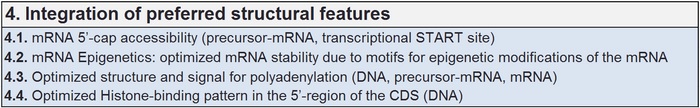
Integrating preferred structural features
Structure functional relations are in general extremely important in Protein and RNA interactions. These relations are obvious and not relative easily to understand. Its correct and functional realizations are still a formidable task which needs sophisticated tools.
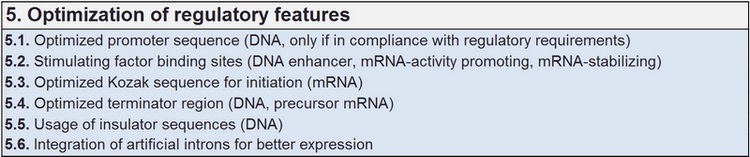
Optimizing regulatory features
Regulatory features are mostly determined by highly specific signal structures, even when such structures are degenerated in the sequence and mostly dependent on positional sequence context. Descriptive motif libraries are deployed for handling molecular function dependent on specific interaction with more or less specific motifs with variable activity.
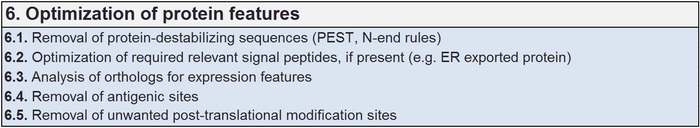
Optimizing protein features
There is a long list of protein features that can support the generation of an improved functional product. Correct folding of proteins has a high impact on the final functional properties of the gene products, including specific activity, transport, efficient cellular traffic, export and correct location. Local amino acid composition, slow codon pattern, protein modification pattern like glycosylation, signal structures, etc., can also have a substantial impact on the protein function.
Resources
Click on ATG Eukaryotic Gene Expression Modulation via BioDesign for more information.
Click on ATG to contact the company directly.
Supplier Information
Supplier: ATG:biosynthetics GmbH
Address: Weberstrasse 40, 79249 Merzhausen, Germany
Tel: +49 (0) 761 888 9424
Fax: +49 761 888 9425
Website: www.atg-biosynthetics.com/