By ATG:biosynthetics GmbH
Gene synthesis of Artificial Genes, it’s Functional Variants and Gene Clusters
Overview
Are you still performing complex molecular cloning work in your lab by yourself? Let‘s use a simple and increasingly popular alternate method in synthetic biology to create gene fragments, functional synthetic genes and higher order gene assemblies thereof.
ATG:biosynthetics is specialist for all types of gene sequence modulations with functional impact and can deliver these gene sequences physically as a valid alternative to PCR amplification and cloning of naturally occurring genes.
Synthetic genes are an attractive approach for those wishing to string together genes or parts of genes, potentially saving repeated rounds of amplification, restriction, ligation, selection, etc.
Scope of gene synthesis
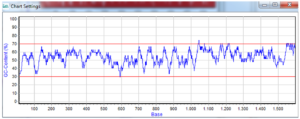
Standard Gene synthesis covers ‘plain vanilla’ DNA sequences that have an even guanine-cytosine (GC) content ranging from 30 to 75 per cent (Figure 1) with no problematic secondary structures, extended repeats or polynucleotide stretches, etc. These ‘run-of-the mill’ genes need no sophisticated and enhanced protocols but can be synthesized with established and time-tested procedures that save time and money.
This is in contrast to complex genes that usually require significant modifications to existing protocols or completely new protocols.
Optimizing auto- and heterologous genes
Even standard genes can be optimized, either by adapting codon usage to another organism or by tweaking the original gene, e.g. by optimizing control regions, streamlining GC content, etc.
ATG’s proprietary EvoMAG (Evolution Mediated Artificial Genetics) software package provides a powerful tool for this, allowing parallel processing of multiple genes and gene variant libraries from different genetic sources. This calculates highly accessible mRNA-structures around the translational initiation regions (TIRs).
EvoMAG can be used to adapt codon use, using its smart ACDC-SD design to optimize assembly cloning, disassembly cloning and substitution design.
Codons selection for to adaptiveness with locally preferred codon use along the message as well as cut-off thresholds for codon usage, with the software set to generate a modified (normalized) usage table (Figure 2).
This offers powerful advantages including adaptability to all genetic systems and molecular approaches, easier reconstruction from ACCEPTOR to DONOR vectors and no theoretical limit in assembly of building blocks.
Resources
Click on Standard gene synthesis for more information.
Click on ATG to contact the company directly.
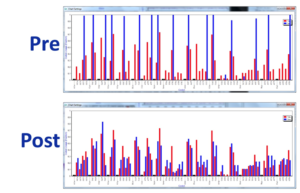
Figure 2: Adapting codon use with EvoMAG showing tabulated frequencies (blue bars) and frequency distribution (red) in the submitted sequence pre-optimization and the optimized sequence (lower image).Not all possible codons are used as a cut-off threshold for codon usage and post- optimization the frequency distribution follows the normalized table more closely.
Supplier Information
Supplier: ATG:biosynthetics GmbH
Address: Weberstrasse 40, 79249 Merzhausen, Germany
Tel: +49 (0) 761 888 9424
Fax: +49 761 888 9425
Website: www.atg-biosynthetics.com/