By Phage Consultants
Phage Consultants guide to bacteriophages
Phage Consultants (PC) provides some of the world’s most expert testing services to detect bacteriophage and prophage presence across a wide spectrum of bacterial hosts.
The company was created to help clients whose production is based on bacterial activities in order to combat bacteriophage infection but also studies phages’ considerable potential to assist medical therapies, particularly in the field of antibiotic resistance.
PC has now prepared a guide to this complex and fascinating world of bacteriophages to help define phages and their possibilities.
What are bacteriophages?
Bacteriophages, informally known as ‘phages’, are ubiquitous viruses that are found wherever bacteria exist and the most common and diverse entities in the biosphere with an estimated 1031 bacteriophages on Earth – more than the combined total for every other organism, including bacteria.
The term literally means ‘devourer of bacteria’ in reference to their essential mechanism of replicating within bacteria and archaea to subvert or destroy them.
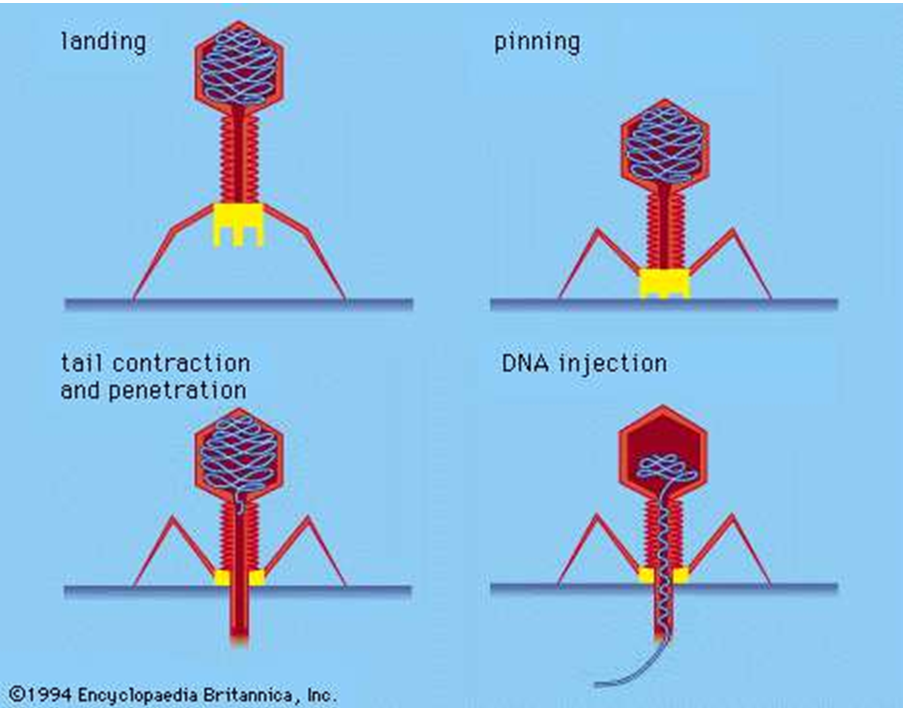
A phage is composed of proteins that encapsulate a DNA or RNA genome, and may have genomic structures encoding as few as four genes, such as the MS2 bacteriophage, up to hundreds of genes. Phages replicate using their ‘alien’ like structure to land on the surface of the bacterium, and inject their genome into the cytoplasm of the bacterium.
A short history of phage research
The existence of bacteriophages has only been known for more than a century, since 1896, when British scientist Ernest Hanbury Hankin reported that something in the waters of the Ganges and Yamuna rivers in India had a marked antibacterial action against cholera and could pass through a very fine porcelain filter.
Formal discovery came during World War 1, when British bacteriologist Frederick Twort discovered in 1915 a small agent that infected and killed bacteria, which he suggested might be a virus that grew on the bacteria and destroyed them, while French-Canadian microbiologist Félix d’Hérelle, working independently at the Pasteur Institute in Paris, announced in September 1917 that he had discovered “an invisible, antagonistic microbe… a virus parasitic on bacteria”, giving it the name bacteriophage.
D’Hérelle observed the case of a dysentery patient cured by administering a ‘phage cocktail’ and thus introduced the concept of phage therapy. This cause was taken up particularly enthusiastically in the Soviet Union and Eastern Europe following WW1, with Giorgi Eliava joining d’Hérelle to carry out pioneering work in the Republic of Georgia during the 1920s and 1930s in using phages to treat bacterial infections, leading on to further studies all the way through the 1950s and 1960s.
However, this work was not echoed in the West, principally because of geopolitical tensions and also because the discovery of penicillin and the consequent antibiotics revolution seemed to make phage therapy irrelevant.
Increasing focus on genomics and viruses from the 1960s made the study of phages more relevant and in 1969, Max Delbrück, Alfred Hershey, and Salvador Luria were awarded the Nobel Prize in Physiology or Medicine for their discoveries of the replication of viruses and their genetic structures, in which bacteriophages were involved.
Increased understanding of the genetic interactions between phages and proteins led on to the evolution of the phage display technique, first described in 1985 George P. Smith in 1985, in which filamentous and other phages are used to connect proteins with the genetic information that encodes them, by causing the phage to “display” a protein of interest outside while containing the gene for the protein inside. These displaying phages can then be screened against other proteins, peptides or DNA sequences, allowing large protein and peptide libraries to be assembled through in vitro selection and ‘panning’ techniques.
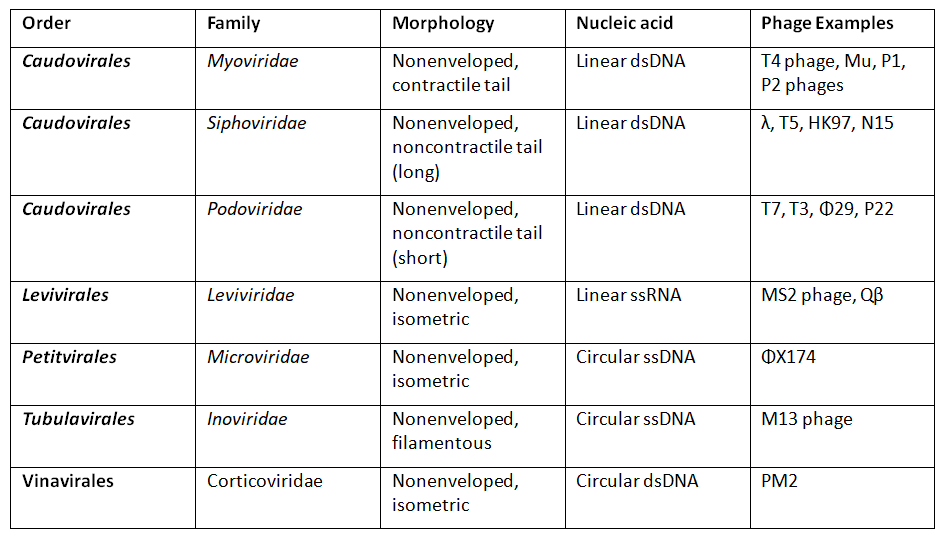
Principal types of bacteriophage
The International Committee on Taxonomy of Viruses (ICTV) classifies bacteriophages according to their morphology and nucleic acid. The principal categories include:
Phage mechanisms
Bacteriophages have three distinct lifecycle models or mechanisms of action:
- Temperate phages: Also called lysogenic phages, these are bacteriophages that carry DNA, which can act as an episome within bacteria. Temperate phages can choose between the lytic (virulent) pathway of development or a lysogenic pathway in which the virus remains dormant until induction, creating a prophage bacterial cell. Temperate bacteriophages start their life cycle when they adsorb to permissive host. After injecting their genome into the host cell, they produce a set of early proteins and a few copies of their genome. Where there are poor growth conditions in the host cell, the phage tends to follow a lysogenic pathway in which it integrates its genetic material with the host cell, ether by physical incorporation into the host genome or by integrating its prophage as a stably maintained plasmid. When a prophage is induced, it starts to produce viral proteins and copies of the viral genome using bacterial resources and biosynthetic apparatus. Progeny virus particles are formed and, after completing the cycle, released during host cell lysis. (see Resources for illustration).
- Filamentous phages: These bacteriophages may cause chronic infections that do not result in cell lysis but rather produce infected bacterial cells that constantly release progeny phage particles. This usually leads to a growth inhibition and a massive, continuous phage production (see Resources for illustration). These characteristics make fd phages useful for phage display research.
- Virulent phages: These start their life cycle when they adsorb to a permissive host, injecting their genetic material into the host and start producing viral proteins and copies of the viral genome using bacterial resources and biosynthetic apparatus. Progeny viral particles are formed and are released after host cell lysis. (see Resources for illustration):
Phages of interest
- Coliphage: A coliphage is a type of bacteriophage that infects Escherichia coli. Examples include bacteriophage lambda and Leviviridae.
- T4 phage: This bacteriophage infects coli bacteria. It is a double-stranded DNA virus that undergoes a lytic lifecycle only T4 is one of the ‘T-even’ strains that also include Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6, all highly complex with genomes made up of around 160 genes and featuring an unusual base hydroxymethylcytosine (HMC) in place of the nucleic acid base cytosine.
- P1 phage: This temperate virus is similar in structure to the T-even phages but simpler, with an icosahedral head[2] containing the genome attached at one vertex to the tail. The tail has a tube surrounded by a contractile sheath. It ends in a base plate with six tail fibres. The tail fibres are involved in attaching to the host and providing specificity. P1 infects coli and some other bacteria and is interesting in research for its potential to transfer DNA from one bacterial cell to another by transduction. P1 can also be used to create an artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.
- M13 phage: M13 is a filamentous bacteriophage composed of circular single-stranded DNA (ssDNA) which is 6407 nucleotides long encapsulated in approximately 2700 copies of the major coat protein P8, and capped with 5 copies of two different minor coat proteins (P9, P6, P3) on the ends. The life cycle of M13 is relatively short, with the early phage progeny exiting the cell ten minutes after infection. M13 is a chronic phage, releasing its progeny without killing the host cells. M13 plasmids are used for many recombinant DNA processes, and the virus has also been used for phage display, directed evolution, nanostructures and nanotechnology applications. M13 is also particularly useful in phage display techniques, along with filamentous phages.
- MS2 phage: Escherichia virus MS2 is an icosahedral, positive-sense single-stranded RNA virus that infects the bacterium Escherichia coli and other members of the Enterobacteriaceae. MS2 is a member of a family of closely related bacterial viruses that includes bacteriophages f2, Qβ, R17, and GA. MS2 was first isolated by Alvin John Clark in 1961 and in 1976, became the first completely sequenced genome, consisting of just 3569 nucleotides of single-stranded RNA encoding only four proteins.
Resources
Click on Temperate phage infection to see animation illustrating mechanism of action.
Click on Filamentous phage to see animation illustrating mechanism of action.
Click on Virulent phage infection to see animation illustrating mechanism of action.